We provide effective and economically affordable training courses for R and Python, Click here for more details and course registration !
- Introduction of t-distribution and t-test
A Student t-distributed random variable is modeling the ratio between a standard Normal random variate and square root of a Chi-squared random variable divided by its degrees of freedom.
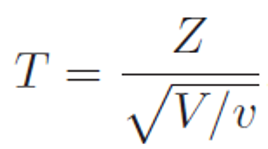
Where
Z is a random variable that follows standard Normal distribution,
and V is a random variable that follows a Chi-squared distribution with degrees of freedom v.
The probability density function for t-distribution is given as
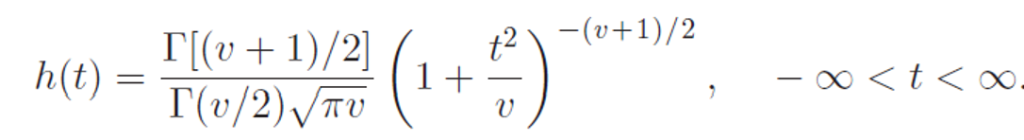
Where
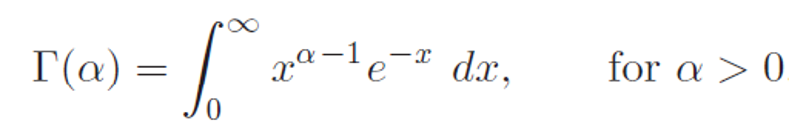
denotes a gamma function.
t-distribution is often used to test a population mean under a Null hypothesis (H0) : μ = μ0 , against an Alternative hypothesis (H1): μ not equal to μ0, with random samples in small size (e.g. < 30) from an approximately normally distributed population.
Let X1,X2, . . . , Xn be n independent random numbers coming from a normal population with mean μ and standard deviation σ. Let sample average and sample variance computed as following
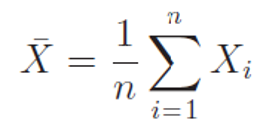
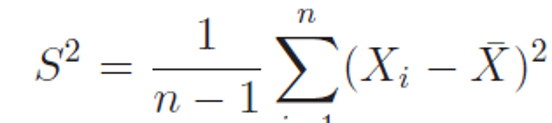
then the test statistics
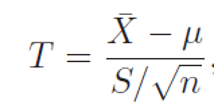
follows a t-distribution with v = n − 1 degrees of freedom. If the T is located at in the critical region, then the test will come to a conclusion of rejecting H0 in favor of H1, or say the test is significant at level α.
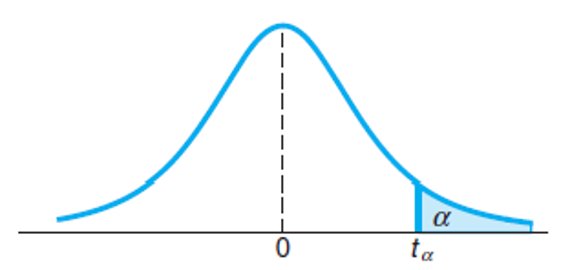
2. Using t-distribution with R
In R programming, there are several functions dealing with t-distribution:
dt(): Probability density
pt(): Cumulative probability
qt(): Quantile value
rt(): Random number generation
dt() function is used to find the value of probability density (pdf) of a random variable x from t-distribution. The basic form of the function is
dt(x, df)
where x is usually a vector represents a series of random variables, and df is the degrees of freedom. Following code example shows how to calculate and plot probability densities for random variables between 0 and 5 from a t-distribution with degrees of freedom 7.
#create a vector between 0 and 5
x_vec <- seq(0, 5, by = 0.1)
df <- 7
#dt() calculating probability density
plot(x_vec, dt(x_vec,df))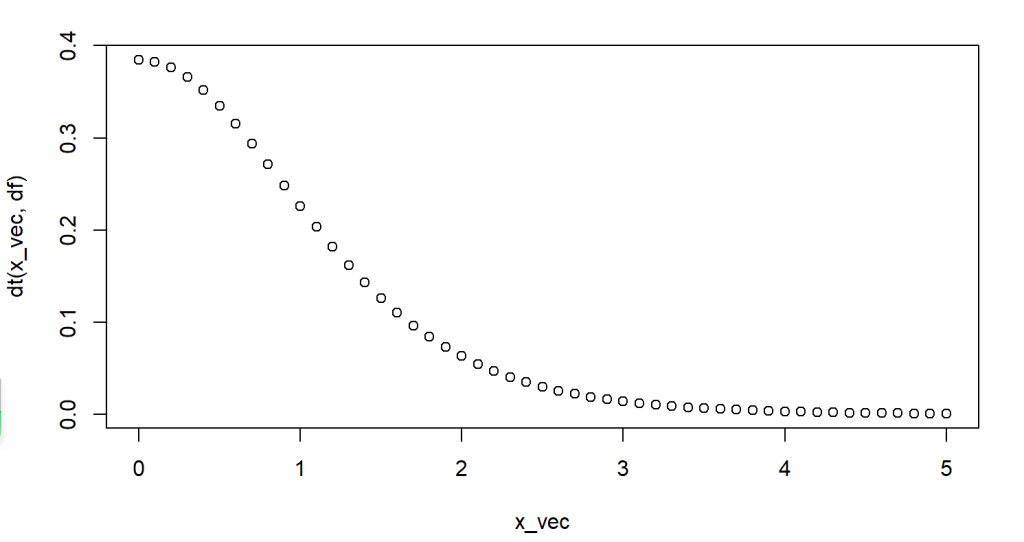
pt() function is used to calculate the cumulative probability (CDF) till a given value following t-distribution. The basic form of the function is
pt(q, df)
where
q represents variates from t-distribution, and df denotes degrees of freedom.
The following code example show calculating and plotting cumulative probabilities for variates between 0 and 5, which follow a t-distribution with degrees of freedom 7. And from the figure we can see that about 80% of the values are less than or equal to 1 in this distribution.
#create a vector of random variables
x_vec <- seq(0, 5, by = 0.1)
df <- 7
#pt() function for calculating cumulative probabilities
#from t-distribution
plot(x_vec, pt(x_vec,df))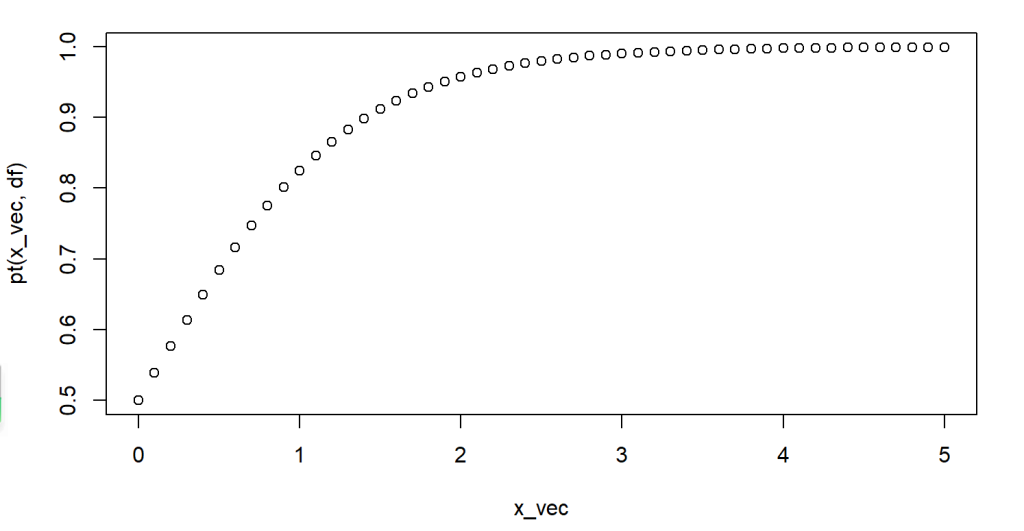
function qt() is the opposite operation of pt(), and is used to calculate the t-distribution quantile value, from the given cumulative probability. The basic form of the function is
qt(p, df)
where p represents the cumulative probability, and df is for degrees of freedom.
The following code example shows calculating and plotting the quantile values for the given cumulative probabilities between 0 and 1, for a t-distribution of degrees of freedom 7. From the figure we can also see that about 80% of the values in the given distribution have values below 1.
#create a vector representing cumulative probabilities
probs <- seq(0, 1, by = 0.05)
df <- 7
#qt() for calculating t-distribution quantile values
plot(probs, qt(probs,df))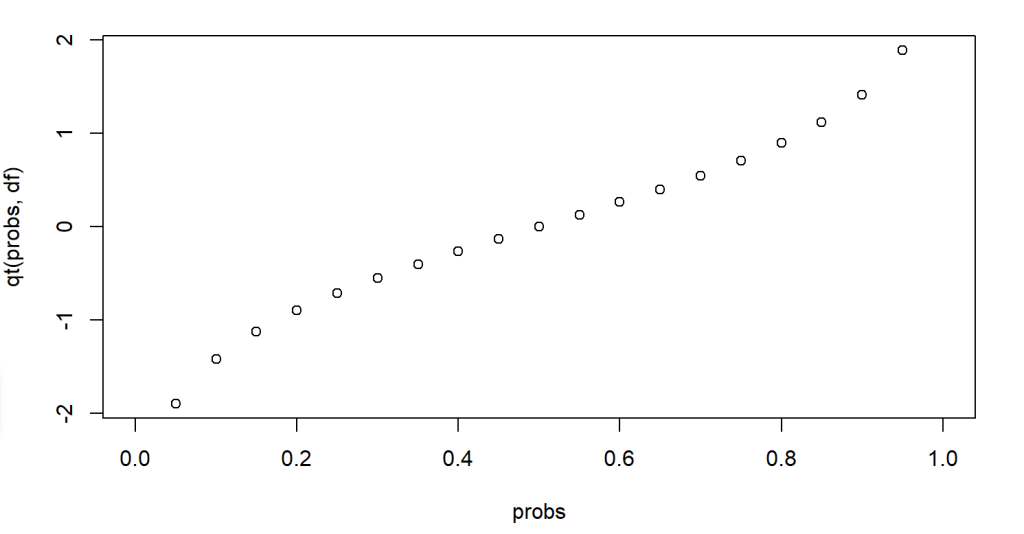
rt() is used to generate random variates from a given t-distribution. The basic syntax of the function is
rt(N, df)
where N is the number of values to generate, and df represents degrees of freedom.
Following code example show generating 10000 random values from a t-distribution of degrees of freedom 10.
# set how many values to generate
N <- 10000
df <- 10
#rt() for t-distribution variates generation
plot(hist(rt(N,df)))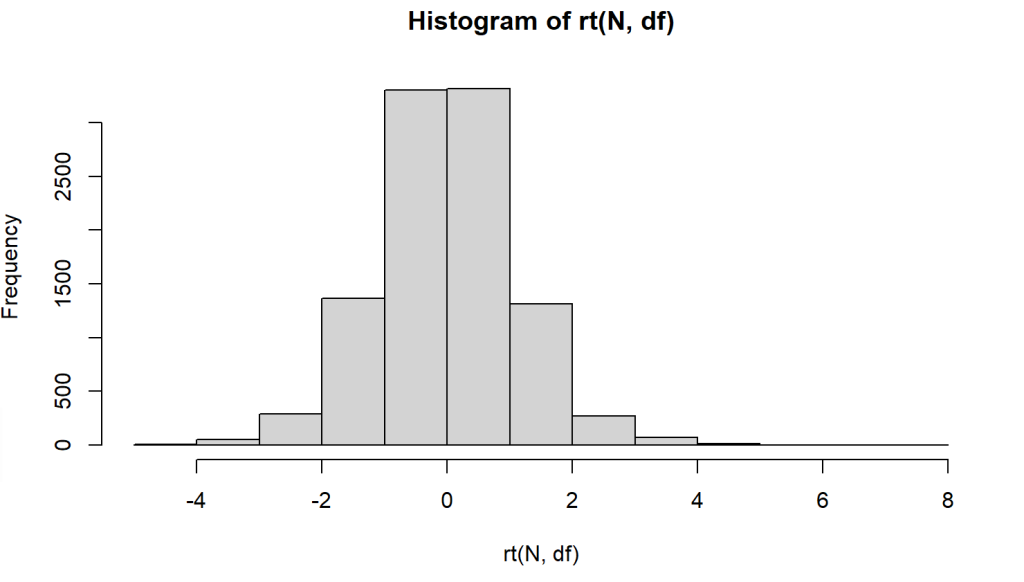
3. Using t-test with R
The following example shows how to use t-distribution functions for t-test in R.
Description of the example:
A medical researcher claims that the population mean yield of a certain medical product is 500 cm. To check this claim he samples 25 batches per week. If the computed t-value falls between −t0.05 and t0.05, he will hold the claim. What conclusion should he draw from a sample that has a mean X-Ba = 518 cm and a sample standard deviation s = 40 cm ? Assume the distribution of the population to be approximately normal.
Solution:
# first we calculate the critical value of t0.05
# using qt(probs, df)
t_cri <- qt( 1-0.05, 24)
t_cri
[1] 1.710882
# then we calculate the sample t test statistics
t_sample <- (518 - 500) / (40/sqrt(25))
t_sample
[1] 2.25
#we can also calcualte the p_value assocated with t_sample
p_sample <- 1 - pt(t_sample, 24)
p_sample
[1] 0.01694426As t_sample > t_cri, it also shows p_sample < 0.05, the medical researcher is likely to reject the claim and conclude that the process yields is probably larger than 500 cm.
You can also watch full video on R tutorial from our YouTube channel.
0 Comments